Fundamentos de la epidemiología genómica, lecciones aprendidas de la enfermedad por coronavirus (COVID-19) y nuevas direcciones
DOI:
https://doi.org/10.54502/msuceva.v2n2a5Palabras clave:
COVID-19, epidemiología molecular, secuenciación genómica, vigilancia genómica mejorada, virulenciaResumen
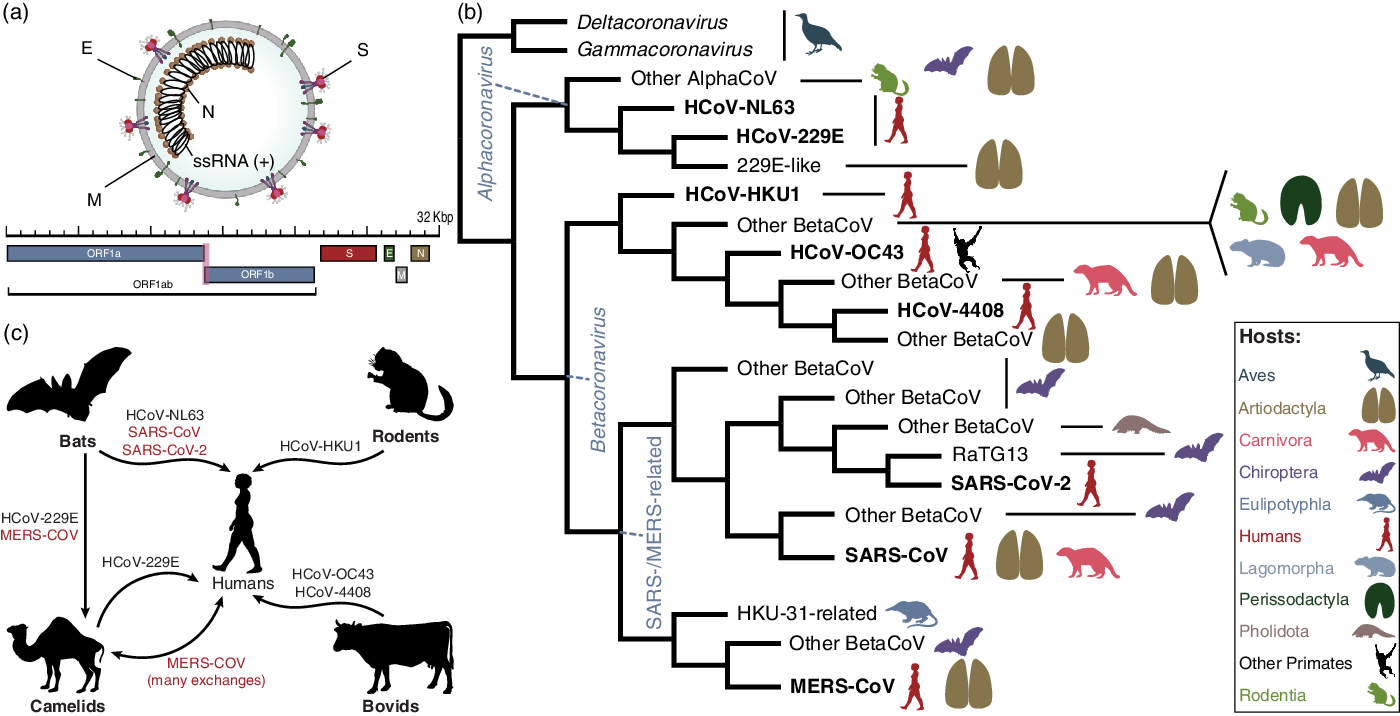
La pandemia de la enfermedad por coronavirus 2019 (COVID-19) fue una de las principales causas de muerte en todo el mundo en 2020. La enfermedad es causada por el coronavirus 2 (SARS-CoV-2), un virus de ARN de la subfamilia Orthocoronavirinae relacionado con otros 2 coronavirus clínicamente relevantes, SARS-CoV y MERS-CoV. Al igual que otros coronavirus y varios otros virus, el SARS-CoV-2 se originó en los murciélagos. Sin embargo, a diferencia de otros coronavirus, el SARS-CoV-2 resultó en una pandemia devastadora. La pandemia de SARS-CoV-2 continúa, debido a la evolución viral que conduce a variantes más transmisibles e inmunes evasivas. Tecnologías como la secuenciación genómica, ha impulsado el cambio de la epidemiología sindrómica a la molecular, y promete una mejor comprensión de las variantes. La pandemia de COVID-19 ha expuesto obstáculos críticos que deben abordarse para desarrollar la ciencia de las pandemias. Gran parte del progreso se está aplicando en el mundo desarrollado.
Sin embargo, persisten las barreras para el uso de la epidemiología molecular en los países de ingresos bajos y medianos (LMIC), incluida la falta de logística para equipos y reactivos y la falta de capacitación en análisis. Revisamos la literatura de epidemiología molecular para comprender sus orígenes desde la epidemia de SARS (2002-2003) hasta los eventos de influenza y la pandemia actual de COVID-19. Abogamos por una mejor vigilancia genómica del SARS-CoV y por comprender la diversidad de patógenos en posibles huéspedes zoonóticos. Este trabajo requerirá capacitación en computación filogenética y de alto rendimiento para mejorar los análisis del origen y la propagación de patógenos. Los objetivos generales son comprender y reducir el riesgo de zoonosis a través de la colaboración interdisciplinaria y la reducción de las barreras logísticas.
Descargas
Métricas
Citas
Janies DA. Phylogenetic concepts and tools applied to epidemiologic investigations of infectious diseases. Microbiol Spectr 2019;7. https://doi.org/10. 1128/microbiolspec.AME-0006-2018
Janies DA, Pomeroy LW, Aaronson JM, et al. Analysis and visualization of H7 influenza using genomic, evolutionary and geographic information in a modular web service. Cladistics 2012;28:483–488. https://doi.org/10.1111/j.1096-0031.2012.00401.x
Hoffmann M, Luo Y, Monday SR, et al. Tracing origins of the Salmonella Bareilly strain causing a foodborne outbreak in the United States. J Infect Dis 2016;213:502–508.
https://doi.org/10.1093/infdis/jiv297
Ezeoke I, Galac MR, Lin Y, et al. Tracking a serial killer: integrating phylo- genetic relationships, epidemiology, and geography for two invasive meningococcal disease outbreaks. PLoS One 2018;13: e0202615. https://doi.org/10.1371/journal.pone.0202615
Allard MW, Strain E, Melka D, et al. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 2016;54: 1975–1983. https://doi.org/ 10.1128/JCM.00081-16
Marra MA, Jones SJM, Astell CR, et al. The genome sequence of the SARS- associated coronavirus. Science 2003;300:1399–1404.
https://doi.org/10.1126/science.1085953
Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003;300:1394–1399.
https://doi.org/10.1126/science.1085952
Boulos MNK. Descriptive review of geographic mapping of severe acute respiratory syndrome (SARS) on the Internet. Int J Health Geogr 2004;3:2. https://doi.org/10.1186/1476-072X-3-2
Janies DA, Habib F, Alexandrov B, Hill A, Pol D. Evolution of genomes, host shifts and the geographic spread of SARS-CoV and related coronaviruses. Cladistics 2008;24:111–130.
https://doi.org/10.1111/j.1096-0031.2008.00199.x
Janies DA, Hill AW, Guralnick R, Habib F, Waltari E, Wheeler WC. Genomic analysis and geographic visualization of the spread of avian influenza (H5N1). Syst Biol 2007;56:321–329. https://doi.org/10.1080/10635150701266848
Janies DA, Treseder T, Alexandrov B, et al. The Supramap project: linking pathogen genomes with geography to fight emergent infectious diseases. Cladistics 2011;27:61–66.
https://doi.org/10.1111/j.1096-0031.2010.00314.x
Studer J, Janies DA. Global spread and evolution of viral haemorrhagic septicaemia virus. J Fish Dis 2011;34:741–747. https://doi.org/10.1111/j.1365-2761.2011.01290.x
Janies DA, Voronkin IO, Studer J, et al. Selection for resistance to oselta- mivir in seasonal and pandemic H1N1 influenza and widespread co-cir- culation of the lineages. Int J Health Geogr 2010;9:13. https://doi.org/10.1186/1476-072X-9-13
Hill AW, Guralnick RP, Wilson MJC, Habib F, Janies D. Evolution of drug resistance in multiple distinct lineages of H5N1 avian influenza. Infect Genet Evol 2009;9:169–178. https://doi.org/10.1016/j.meegid.2008.10.006
Janies, DA. Local challenges of global proportions: evaluating role, preparedness for, and surveillance for pandemic influenza: Hearing before the committee on homeland security and government affairs, United States senate, 1 Sess. (2007). US government website. https://www.govinfo.gov/content/pkg/CHRG-110shrg38846/ html/CHRG-110shrg38846.htm
Janies DA, Voronkin IO, Das M, Hardman J, Treseder TW, Studer J. Genome informatics of influenza A: from data sharing to shared analytical capabilities. Anim Health Res Rev 2010;11:73–79. https://doi.org/10.1017/S1466252310000083
Ruan YJ, Wei CL, Ee AL, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 2003;361:1779–1785. https://doi.org/10.1016/s0140-6736(03)13414-9
Zhao G-P. SARS molecular epidemiology: a Chinese fairy tale of control- ling an emerging zoonotic disease in the genomics era. Philos Trans R Soc Lond B Biol Sci 2007;362:1063–1081. https://doi.org/ 10.1098/rstb.2007.2034
Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell 2015;58:586–597.
https://doi.org/10.1016/j.molcel.2015.05.004
Pérez-Losada M, Arenas M, Galán JC, et al. High-throughput sequencing (HTS) for the analysis of viral populations. Infect Genet Evol 2020;80:104208. https://doi.org/10.1016/j.meegid.2020.104208
National Human Genome Research Institute. The Cost of Sequencing a Human Genome, 2021. https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost
Woo PCY, Lau SKP, Lam CSF, et al. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal corona virus in gamma coronavirus. J Virol 2014;88:1318–1331. https://doi.org/10.1128/JVI.02351-13
Durães-Carvalho R, Caserta LC, Barnabé ACS, et al. Coronaviruses detected in Brazilian wild birds reveal close evolutionary relationships with beta- and deltacoronaviruses isolated from mammals. J Mol Evol 2015;81:21–23.
https://doi.org/10.1007/s00239-015-9693-9
Woo PCY, Lau SKP, Lam CSF, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coro naviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and delta- coronavirus. J Virol 2012;86:3995–4008. https://doi.org/10.1128/JVI.06540-11
Woo PCY, Huang Y, Lau SKP, Yuen K-Y. Coronavirus genomics and bio- informatics analysis. Viruses. 2010;2:1804–1820. https://doi.org/10.3390/v2081803
Irigoyen N, Firth AE, Jones JD, Chung BY-W, Siddell SG, Brierley I. High- resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog 2016;12:e1005473. https://doi.org/10.1371/journal.ppat.1005473
ViralZone. Coronavirinae; 2021. https://viralzone.expasy.org/785? outline=all_by_species
Li BX, Ge JW, Li YJ. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. https://doi.org/10.1016/j.virol.2007.03.031
Boileau MJ, Kapil S. Bovine coronavirus associated syndromes. Vet Clin N Am Food Anim Pract 2010;26:123–146. https://doi.org/10.1016/j.cvfa.2009.10.003
Mandelik R, Sarvas M, Jackova A, Salamunova S, Novotny J, Vilcek S. First outbreak with chimeric swine enteric coronavirus (SeCoV) on pig farms in Slovakia—lessons to learn. Acta Vet Hung 2018;66:488–492. https://doi.org/10.1556/004.2018.043
Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016;24:490–502. https://doi.org/10.1016/j.tim.2016.03.003
Lai MMC, Cavanagh D. The molecular biology of coronaviruses. In Advances in Virus Research. New York: Elsevier; 1997: 1–100. https://doi.org/10.1016/S0065-3527(08)60286-9
Centers for Disease Control and Prevention. Human coronavirus types; 2021, March 17.
https://www.cdc.gov/coronavirus/types.html.
Zhang XM, Herbst W, Kousoulas KG, Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J Med Virol 1994;44:152–161. https://doi.org/10.1002/jmv.1890440207
Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February 2003. Lancet 2003;362:1353–1358. https://doi.org/10.1016/s0140-6736(03)14630-2
Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953–1966. https://doi.org/ 10.1056/NEJMoa030781
Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet 2015;386:995–1007.
https://doi.org/10.1016/S0140-6736(15)60454-8
World Health Organization. World Health Organization director-general’s opening remarks at the media briefing on COVID-19. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-11-march-2020.
Pekar J, Worobey M, Moshiri N, Scheffler K, Wertheim JO. Timing the SARS-CoV-2 index case in Hubei province. Science 2021;372:412–417. https://doi.org/10.1126/science.abf8003
Coronavirus (COVID-19) dashboard. World Health Organization. https://covid19.who.int.
Yuen K-S, Ye Z-W, Fung S-Y, Chan C-P, Jin D-Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci 2020;10:40. https://doi.org/10.1186/s13578-020-00404-4
Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proxi mal origin of SARS-CoV-2. Nat Med 2020;26:450–452. https://doi.org/10.1038/s41591-020-0820-9
Liu S-L, Saif LJ, Weiss SR, Su L. No credible evidence supporting claims of the laboratory engineering of SARS-CoV-2. Emerg Microbes Infect 2020;9:505–507. https://doi.org/10.1080/22221751.2020.1733440
Holmes EC, Goldstein SA, Rasmussen AL, et al. The origins of SARS- CoV-2: a critical review. Cell 2021, 184(19):4848-4856. https://doi.org/10.1016/j.cell.2021.08.017
Wang N, Li S-Y, Yang X-L, et al. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol Sin 2018;33:104–107. https://doi.org/10.1007/s12250-018-0012-7
Zhang X, Hasoksuz M, Spiro D, et al. Quasi-species of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology 2007;363:1–10. https://doi.org/10.1016/j.virol.2007.03.018
Rasmussen AL. On the origins of SARS-CoV-2. Nat Med, 27,9 (2021). https://doi.org/10.1038/s41591-020-01205-5
Shi Z-L. Origins of SARS-CoV-2: focusing on science. Infect Dis Immun 2021;1:3–4.
https://doi.org/10.1097/ID9.0000000000000008
Machado DJ, Scott R, Guirales S, Janies DA. Fundamental evolution of all including three deadly lineages descendent from Chiroptera-hosted coro naviruses: SARS-CoV, MERS-CoV, and SARS-CoV-2. Cladistics 2021, 27(5), 461-488. https://doi.org/10.1111/cla.12454
Zhao S, Zhuang Z, Cao P, et al. Quantifying the association between domestic travel and the exportation of novel coronavirus (2019-nCoV) cases from Wuhan, China in 2020: a correlational analysis. J Travel Med 2020;27. https://doi.org/10.1093/jtm/taaa022
Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005;310:676–679.
https://doi.org/10.1126/science.1118391
Lytras S, Hughes J, Martin D, et al. Exploring the natural origins of SARS- CoV-2 in the light of recombination. bioRxiv 2021. https://doi.org/10.1101/2021.01. 22.427830
Lytras S, Xia W, Hughes J, Jiang X, Robertson DL. The animal origin of SARS-CoV-2. Science 2021;373:968–970. https://doi.org/10.1126/science.abh0117
Lam TT-Y, Jia N, Zhang Y-W, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020;583:282–285. https://doi.org/10.1038/s41586-020-2169-0
Xiao K, Zhai J, Feng Y, et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020;583:286–289. https://doi.org/10.1038/s41586-020-2313-x
Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol 2020;30:1346–1351.e2. https://doi.org/10.1016/j.cub.2020.03.022
Liu P, Jiang J-Z, Wan X-F, et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog 2020;16:e1008421. https://doi.org/10.1371/journal.ppat.1008421
GISAID—Initiative. Global Initiative on Sharing All Influenza Data website. https://www.gisaid.org
GISAID—Initiative. EpiCov website. https://www.epicov.org/
Wu F, Zhao S, Yu B, et al. Author correction: a new coronavirus associated with human respiratory disease in China. Nature 2020;580:E7. https://doi.org/10.1038/s41586-020-2008-3
European Centre for Disease Control and Prevention.
Sequencing of SARS-CoV-2.
https://www.ecdc.europa.eu/sites/default/files/docu ments/sequencing-of-SARS-CoV-2.pdf
World Health Organization. Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. https://www.who.int/publications/i/item/9789240018440
Truelove S, Smith CP, Qin M, et al. Projected resurgence of COVID-19 in the United States in July–December 2021 resulting from the increased transmissibility of the Delta variant and faltering vaccination. medRxiv 2021.
https://doi.org/10.1101/2021.08.28.21262748
Lau BT, Pavlichin D, Hooker AC, et al. Profiling SARS-CoV-2 mutation fingerprints that range from the viral pangenome to individual infection quasispecies. Genome Med 2021;13:62. https://doi.org/10.1186/s13073-021-00882-2
Hamza M, Ali A, Khan S, et al. nCOV-19 peptides mass fingerprinting identification, binding, and blocking of inhibitors flavonoids and anthraquinone of and hydroxychloroquine. J Biomol Struct Dyn 2021;39:4089– 4099.
https://doi.org/10.1080/07391102.2020.1778534
Hariton E, Locascio JJ. Randomised controlled trials—the gold standard for effectiveness research: Study design: randomised controlled trials. BJOG 2018;125:1716.
https://doi.org/10.1111/1471-0528.15199
Boulware DR, Pullen MF, Bangdiwala AS, et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for COVID-19. N Engl J Med. 2020;383:517–525. https://doi.org/ 10.1056/NEJMoa2016638
Siemieniuk RA, Bartoszko JJ, Ge L, et al. Drug treatments for COVID-19: living systematic review and network meta-analysis. BMJ 2020;370: m2980. https://doi.org/10.1136/bmj.m2980
National Human Genome Research Institute. COVID-19 mRNA vaccine production. https://www.genome.gov/about-genomics/ fact-sheets/COVID-19-mRNA-Vaccine-Production
Chiara M, D’Erchia AM, Gissi C, et al. Next-generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Brief Bioinform 2021;22:616–630.
https://doi.org/10.1093/bib/bbaa297
Shaman J, Galanti M. Will SARS-CoV-2 become endemic? Science 2020;370:527–529.
https://doi.org/10.1126/science.abe5960
Nakanishi N, Yoshio I. The novel coronavirus pandemic and the state of the epidemic in Kobe, Japan. J Disaster Res 2021;16:84–87.https://www.fujipress.jp/main/wp-content/themes/Fujipress/pdf_subscribed.php
Phillips N. The coronavirus is here to stay—here’s what that means. Nature 2021;590:382–384.
https://doi.org/10.1038/d41586-021-00396-2
Zhou D, Dejnirattisai W, Supasa P, et al. Evidence of escape of SARS- CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021;184:2348–2361.e6. https://doi.org/10.1016/j.cell.2021.02.037
Kustin T, Harel N, Finkel U, et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA- vaccinated individuals. Nat Med 2021;27:1379–1384. https://doi.org/10.1038/s41591-021-01413-7
Farinholt T, Doddapaneni H, Qin X, et al. Transmission event of SARS- CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med 2021;19:255. https://doi.org/ 10.1186/s12916-021-02103-4
Giovanetti M, Benedetti F, Campisi G, et al. Evolution patterns of SARS- CoV-2: snapshot on its genome variants. Biochem Biophys Res Commun 2021;538:88–91. https://doi.org/10.1016/j.bbrc.2020.10.102
To KK-W, Sridhar S, Chiu KH-Y, et al. Lessons learned 1 year after SARS- CoV-2 emergence leading to COVID-19 pandemic. Emerg Microbes Infect 2021;10:507–535. https://doi.org/ 10.1080/22221751.2021.1898291
Munafò MR, Tilling K, Taylor AE, Evans DM, Davey Smith G. Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol 2018;47:226–235. https://doi.org/10.1093/ije/dyx206
Hernán MA. Invited commentary: selection bias without colliders. Am J Epidemiol 2017;185:1048–1050.
https://doi.org/10.1093/aje/kwx077
Tattan-Birch H, Marsden J, West R, Gage SH. Assessing and addressing collider bias in addiction research: the curious case of smoking and COVID-19. Addiction 2021;116:982–984.
https://doi.org/10.1111/add.15348
Brito AF, Semenova E, Dudas G, et al. Global disparities in SARS-CoV-2 genomic surveillance. medRxiv 2021.
https://doi.org/10.1101/2021.08.21.21262393
Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2-what do they mean? JAMA 2021;325:529–531.
https://doi.org/10.1001/jama.2020.27124
Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS- CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.
https://doi.org/10.1101/2020.12.21.20248640
Ford CT, Scott R, Machado DJ, Janies D. Sequencing data of North American SARS-CoV-2 isolates shows widespread complex variants. medRxiv 2021. https://doi.org/10.1101/2021.01.27.21250648
Awadasseid A, Wu Y, Tanaka Y, Zhang W. Current advances in the devel- opment of SARS-CoV-2 vaccines. Int J Biol Sci 2021;17:8–19. https://doi.org/10.7150/ijbs.52569
Hossain MK, Hassanzadeganroudsari M, Apostolopoulos V. The emer- gence of new strains of SARS-CoV-2. What does it mean for COVID- 19 vaccines? Expert Rev Vaccines 2021;20:635–638.
https://doi.org/10.1080/14760584.2021.1915140
Ul-Rahman A, Shabbir MAB, Aziz MW, et al. A comparative phylogenomic analysis of SARS-CoV-2 strains reported from non-human mammalian species and environmental samples. Mol Biol Rep 2020;47: 9207–9217. https://doi.org/10.1007/s11033-020-05879-5
Kuhn JH, Bao Y, Bavari S, et al. Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch Virol 2013;158:301–311. https://doi.org/10.1007/s00705-012-1454-0
Parums D. Editorial: revised World Health Organization (WHO) terminology for variants of concern and variants of interest of SARS-CoV-2. Med Sci Monit 2021;27:e933622. https://doi.org/10.12659/MSM.933622
Konings F, Perkins MD, Kuhn JH, et al. SARS-CoV-2 variants of interest and concern naming scheme conducive for global discourse. Nat Microbiol 2021;6:821–823. https://doi.org/10.1038/s41564-021-00932-w
92. Janik E, Niemcewicz M, Podogrocki M, Majsterek I, Bijak M. The emerging concern and interest SARS-CoV-2 variants. Pathogens 2021;10. https://doi.org/10.3390/pathogens10060633
Tracking SARS-CoV-2 variants. World Health Organization https://www.who.int/activities/tracking-SARS-CoV-2-variants.
SARS-CoV-2 variant classifications and definitions. Centers for Disease Control and Prevention.
https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html.
Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data from vision to reality. Euro Surveill 2017;22. https://doi.org/10.2807/1560-7917. ES.2017.22.13.30494
Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018;34:4121–4123. https://doi.orgb/10.1093/bioinformatics/bty407
Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020;5:1403–1407. https://doi.org/10.1038/s41564-020-0770-5
Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021;19:409–424. https://doi.org/10.1038/s41579-021-00573-0
Flanagan KL, MacIntyre CR, McIntyre PB, Nelson MR. SARS-CoV-2 vac- cines: where are we now? J Allergy Clin Immunol Pract 2021. https://doi.org/10.1016/ j.jaip.2021.07.016
Cevik M, Grubaugh ND, Iwasaki A, Openshaw P. COVID-19 vaccines: keeping pace with SARS-CoV-2 variants. Cell 2021. https://doi.org/10.1016/j.cell. 2021.09.010
Schmitz AJ, Turner JS, Liu Z, et al. A vaccine-induced public antibody protects against SARS-CoV-2 and emerging variants. Immunity 2021;54: 2159–2166. https://doi.org/10.1016/j.immuni.2021.08.013
Boehm E, Kronig I, Neher RA, et al. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect 2021;27: 1109–1117. https://doi.org/ 10.1016/j.cmi.2021.05.022
Farooqi T, Malik JA, Mulla AH, et al. An overview of SARS-COV-2 epidemiology, mutant variants, vaccines, and management strategies. J Infect Public Health 2021. https://doi.org/10.1016/j.jiph.2021.08.014
Krause PR, Gruber MF. Emergency use authorization of COVID vaccines safety and efficacy follow-up considerations. N Engl J Med 2020;383: e107. https://doi.org/10.1056/NEJMp2031373
Pascual-Iglesias A, Canton J, Ortega-Prieto AM, Jimenez-Guarden˜o JM, Regla-Nava JA. An overview of vaccines against SARS-CoV-2 in the COVID-19 pandemic era. Pathogens 2021;10.
https://doi.org/10.3390/patho gens10081030
Chen Y, Zhu L, Huang W, et al. Potent RBD-specific neutralizing rabbit monoclonal antibodies recognize emerging SARS-CoV-2 variants elicited by DNA prime-protein boost vaccination. Emerg Microbes Infect 2021;10: 1390–1403. https://doi.org/10.1080/22221751.2021.1942227
Hodgson SH, Mansatta K, Mallett G, Harris V, Emary KRW, Pollard AJ. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect Dis 2021;21:e26–e35. https://doi.org/ 10.1016/S1473-3099(20)30773-8
Zhao T, Hu C, Ayaz Ahmed M, Cheng C, Chen Y, Sun C. Warnings regarding the potential coronavirus disease 2019 (COVID-19) transmission risk: vaccination is not enough. Infect Control Hosp Epidemiol 2021;2. https://doi.org/10.1016/j.xinn.2021.100116
Lanzavecchia S, Beyer KJ, Evina Bolo S. Vaccination is not enough: understanding the increase in cases of COVID-19 in Chile despite a high vaccination rate. Epidemiologia 2021;2:377–390. https://doi.org/10.3390/epidemiologia2030028
Poudel U, Subedi D, Pantha S, Dhakal S. Animal coronaviruses and coronavirus disease 2019: lesson for One Health approach. Open Vet J 2020;10: 239–251. https://doi.org/10.4314/ovj.v10i3.1
Semenza JC, Menne B. Climate change and infectious diseases in Europe. Lancet Infect Dis 2009;9:365–375.
https://doi.org/10.1016/S1473-3099(09)70104-5
Bartlow AW, Manore C, Xu C, et al. Forecasting zoonotic infectious disease response to climate change: mosquito vectors and a changing environment. Vet Sci China 2019;6. https://doi.org/ 10.3390/vetsci6020040
Mersha C, Tewodros F. One Health, one medicine, one world: cojoint of animal and human medicine with perspectives, a review. Veterinary World 2012, 5(4), 238-243. https://doi.org/10.5455/vetworld.2012.238-243
Sánchez-Vizcaíno JM. One world, One Health, one virology. Vet Microbiol 2013. https://doi.org/10.1016/j.vetmic.2013.02.018
Reeve-Johnson L. One Health and a world of opportunity. Veterinary Record 2015. https://doi.org/10.1136/vr.h2117
Mwangi W, de Figueiredo P, Criscitiello MF. One Health: addressing global challenges at the nexus of human, animal, and environmental Health. PLoS Pathog 2016;12:e1005731.
https://doi.org/10.1371/journal.ppat.1005731
Kelly TR, Karesh WB, Johnson CK, et al. One Health proof of concept: bringing a transdisciplinary approach to surveillance for zoonotic viruses at the human–wild animal interface. Prev Vet Med 2017;137:112–118. https://doi.org/10.1016/j.prevetmed.2016.11.023
Colella JP, Stephens RB, Campbell ML, Kohli BA, Parsons DJ, Mclean BS. The open-specimen movement. Bioscience 2021;71:405–414. https://doi.org/10.1093/biosci/biaa146
Cook JA, Arai S, Armién B, et al. Integrating biodiversity infrastructure into pathogen discovery and mitigation of emerging infectious diseases. Bioscience 2020;70:531–534.
https://doi.org/10.1093/biosci/biaa064
Thompson CW, Phelps KL, Allard MW, et al. Preserve a voucher specimen! The critical need for integrating natural history collections in infectious disease studies. MBio 2021;12.
https://doi.org/10.1128/mBio.02698-20
Bakker FT, Antonelli A, Clarke JA, et al. The Global Museum: natural history collections and the future of evolutionary science and public education. Peer J 2020;8:e8225. https://doi.org/10.7717/peerj.8225
Colella JP, Bates J, Burneo SF, et al. Leveraging natural history biorepositories as a global, decentralized, pathogen surveillance network. PLoS Pathog 2021;17:e1009583. https://doi.org/10.1371/journal.ppat.1009583
Conceicao C, Thakur N, Human S, et al. The SARS-CoV-2 spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol 2020;18: e3001016. https://doi.org/10.1371/journal.pbio.3001016
Wang L, Mitchell PK, Calle PP, et al. Complete genome sequence of SARS-CoV-2 in a tiger from a US zoological collection. Microbiol Resour Announc 2020;9. https://doi.org/10.1128/MRA.00468-20
McAloose D, Laverack M, Wang L, et al. From people to: natural SARS- CoV-2 infection in tigers and lions at the Bronx Zoo. MBio 2020;11. https://doi.org/10.1128/mBio.02220-20
Bartlett SL, Diel DG, Wang L, et al. SARS-CoV-2 infection and longitudinal fecal screening in Malayan tigers (Panthera tigris jacksoni), Amur tigers (Panthera tigris altaica), and African lions (Panthera leo kru- geri) at the Bronx Zoo, New York, USA. J Zoo Wildl Med 2021;51:733–744. https://doi.org/10.1638/2020-0171
Oreshkova N, Molenaar RJ, Vreman S, et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill 2020;25. https://doi.org/10.2807/1560-7917.ES.2020.25.23.2001005
Hammer AS, Quaade ML, Rasmussen TB, et al. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg Infect Dis 2021;27:547–551. https://doi.org/10.3201/eid2702.203794
Halfmann PJ, Hatta M, Chiba S, et al. Transmission of SARS-CoV-2 in domestic cats. N Engl J Med 2020;383:592–594. https://doi.org/10.1056/NEJMc2013400
Gaudreault NN, Trujillo JD, Carossino M, et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg Microbes Infect 2020;9:2322–2332. https://doi.org/ 10.1080/22221751.2020.1833687
Braun KM, Moreno GK, Halfmann PJ, et al. Transmission of SARS-CoV- 2 in domestic cats imposes a narrow bottleneck. PLoS Pathog 2021;17: e1009373. https://doi.org/10.1371/journal.ppat.1009373
Liu H-L, Yeh I-J, Phan NN, et al. Gene signatures of SARS-CoV/SARS- CoV-2–infected ferret lungs in short- and long-term models. Infect Genet Evol 2020;85:104438. https://doi.org/10.1016/j.meegid.2020.104438
Ryan KA, Bewley KR, Fotheringham SA, et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat Commun 2021;12:81. https://doi.org/10.1038/s41467-020-20439-y
Kim Y-I, Kim S-G, Kim S-M, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020;27:704–709.e2. https://doi.org/10.1016/j.chom.2020.03.023
Freuling CM, Breithaupt A, Müller T, et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg Infect Dis 2020;26: 2982–2985. https://doi.org/10.3201/eid2612.203733
Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020;585: 268–272. https://doi.org/10.1038/s41586-020-2324-7
Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID- 19, MERS, and SARS in a nonhuman primate model. Science 2020; 368:1012–1015. https://doi.org/10.1126/science.abb7314
Mykytyn AZ, Lamers MM, Okba NMA, et al. Susceptibility of rabbits to SARS-CoV-2. Emerg Microbes Infect 2021;10:1–7. https://doi.org/10.1080/22221751.2020.1868951
Schlottau K, Rissmann M, Graaf A, et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe 2020;1:e218–e225. https://doi.org/10.1016/S2666-5247(20)30089-6
Imai M, Iwatsuki-Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Nat Acad Sci 2020;117:16587–16595. https://doi.org/10.1073/pnas.2009799117
Palmer MV, Martins M, Falkenberg S, et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J Virol 2021. https://doi.org/10.1128/ JVI.00083-21
Chandler JC, Bevins SN, Ellis JW, et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). bioRxiv 2021. https://doi.org/10.1101/ 2021.07.29.454326
Gryseels S, De Bruyn L, Gyselings R, Calvignac-Spencer S, Leendertz FH, Leirs H. Risk of human-to-wildlife transmission of SARS-CoV-2. Mamm Rev 2020. https://doi.org/10.1111/mam.12225
Griffin JB, Haddix M, Danza P, et al. SARS-CoV-2 Infections and hospitalizations among persons aged ≥16 years, by vaccination status—Los Angeles County, California, May 1–July 25, 2021. Morb Mortal Wkly Rep 2021;70:1170–1176. https://doi.org/ 10.15585/mmwr.mm7034e5
Del Rio C, Malani PN, Omer SB. Confronting the delta variant of SARS-CoV-2, summer 2021. JAMA 2021.
https://doi.org/10.1001/jama.2021. 14811
Lazarevic I, Pravica V, Miljanovic D, Cupic M. Immune evasion of SARS- CoV-2 emerging variants: what have we learnt so far? Viruses 2021;13. https://doi.org/10.3390/v13071192
Kemp SA, Collier DA, Datir RP, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021;592:277–282. https://doi.org/10.1038/s41586-021-03291-y
Srivastava S, Banu S, Singh P, Sowpati DT, Mishra RK. SARS-CoV-2 genomics: an Indian perspective on sequencing viral variants. J Biosci 2021;46. https://doi.org/10.1007/s12038-021-00145-7
Furuse Y. Genomic sequencing effort for SARS-CoV-2 by country during the pandemic. Int J Infect Dis 2021;103:305–307. https://doi.org/10.1016/j.ijid.2020.12.034
Crawford DC, Williams SM. Global variation in sequencing impedes SARS-CoV-2 surveillance. PLoS Genet 2021;17:e1009620. https://doi.org/10.1371/journal.pgen.1009620
Adepoju P. Challenges of SARS-CoV-2 genomic surveillance in Africa. Lancet Microbe 2021;2:e139.
https://doi.org/10.1016/S2666-5247(21)00065-3
Blomberg N, Lauer KB. Connecting data, tools and people across Europe: ELIXIR’s response to the COVID-19 pandemic. Eur J Hum Genet 2020;28:719–723. https://doi.org/10.1038/s41431-020-0637-5
Conesa A, Beck S. Making multiomics data accessible to researchers. Sci Data 2019;6:251.
https://doi.org/10.1038/s41597-019-0258-4
Hodcroft EB, De Maio N, Lanfear R, et al. Want to track pandemic variants faster? Fix the bioinformatics bottleneck. Nature 2021; 591:30–33. https://doi.org/10.1038/d41586-021-00525-x
Turakhia Y, Thornlow B, Hinrichs AS, et al. Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS- CoV-2 pandemic. Nat Genet 2021;53:809–816. https://doi.org/10.1038/s41588-021-00862-7
Wenzel J. Origins of SARS-CoV-1 and SARS-CoV-2 are often poorly explored in leading publications. Cladistics 2020;36:374–379. https://doi.org/10.1111/cla.12425
Machado DJ, Schneider AB, Guirales S, Janies DA. FLAVi: an enhanced annotator for viral genomes of Flaviviridae. Viruses 2020;12. https://doi.org/10.3390/ v12080892
Wheeler WC. Sequence alignment, parameter sensitivity, and the phylogenetic analysis of molecular data. Syst Biol 1995;44:321–331.
Grant T. The perils of “point-and-click” systematics. Cladistics 2003;19: 276–285.
https://doi.org/10.1016/S0748-3007(03)00029-X
Hovmöller R, Alexandrov B, Hardman J, Janies D. Tracking the geographical spread of avian influenza (H5N1) with multiple phylogenetic trees. Cladistics 2010;26:1–13. https://doi.org/10.1111/j.1096-0031.2009.00297.x
de Bernardi Schneider A, Ford CT, et al. StrainHub: a phylogenetic tool to construct pathogen transmission networks. Bioinformatics 2020;36:945–947. https://doi.org/10.1093/bioinformatics/btz646
Syrowatka A, Kuznetsova M, Alsubai A, et al. Leveraging artificial intelligence for pandemic preparedness and response: a scoping review to identify key use cases. NPJ Digit Med 2021;4:96. https://doi.org/10.1038/s41746-021-00459-8
Chen S, Owolabi Y, Li A, et al. Patch dynamics modeling framework from pathogens’ perspective: Unified and standardized approach for complicated epidemic systems. PLoS One 2020;15:e0238186. https://doi.org/10.1371/journal.pone.0238186
COVID-19. Johns Hopkins Coronavirus Resource Center website. https:// coronavirus.jhu.edu/map.html
Henderson DA. The eradication of smallpox—an overview of the past, present, and future. Vaccine 2011;29 suppl 4:D7–9. https://doi.org/10.1016/j.vaccine.2011.06.080
Arunkumar G, Chandni R, Mourya DT, et al. Outbreak investigation of Nipah virus disease in Kerala, India, 2018. J Infect Dis 2019;219: 1867–1878. https://doi.org/10.1093/infdis/jiy612
Soh SM, Kim Y, Kim C, Jang US, Lee H-R. The rapid adaptation of SARS- CoV-2-rise of the variants: transmission and resistance. J Microbiol 2021;59:807–818. https://doi.org/ 10.1007/s12275-021-1348-5
Schwarze K, Buchanan J, Fermont JM, et al. The complete costs of genome sequencing: a microcosting study in cancer and rare diseases from a single center in the United Kingdom. Genet Med 2020;22:85–94. https://doi.org/10.1038/s41436-019-0618-7
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2022 Machado, DJ; White III, RA; Kofsky, J; Janies, DA

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Magna Scientia UCEVA proporciona un acceso abierto, libre y gratuito a su contenido, basado en el principio de que ofrecer al público un acceso libre a las investigaciones, ayuda a un mayor intercambio global del conocimiento. Lo cual, implica que los usuarios pueden leer, descargar, almacenar, imprimir, buscar, indexar y realizar enlaces a los textos completos de esta revista. Se permite distribuir los diversos artículos en las versiones post-print y oficial, sin previo permiso del autor o editor, considerando que el fin de este, no implica fines comerciales, ni la generación de obras derivadas; Solo se solicita la mención de la fuente así como la autoría. El titular del copyright será el o los autores que publiquen en Magna Scientia UCEVA.
Magna Scientia UCEVA está distribuida bajo los términos de la licencia https://creativecommons.org/licenses/by-nc-nd/4.0/deed.es